Transformations#
Before you can start with a second level analysis you are facing the problem that all your output from the first level analysis are still in their subject specific subject-space. Because of the huge differences in brain size and cortical structure, it is very important to transform the data of each subject from its individual subject-space into a common standardized reference-space. This process of transformation is what we call normalization and it consists of a rigid body transformation (translations and rotations) as well as of a affine transformation (zooms and shears). The most common template that subject data is normalized to is the MNI template.
MNI Space and templates
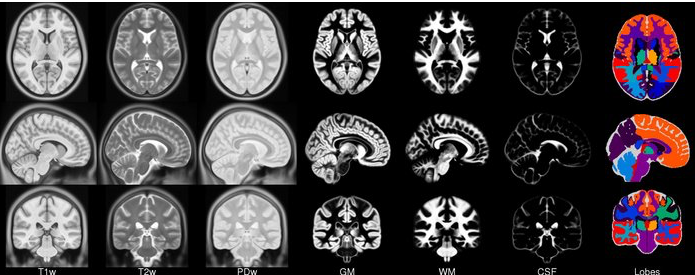
The Montreal Neurological Institute (MNI) has published several “template brains,” which are generic brain shapes created by averaging together hundreds of individual anatomical scans. There are linear and non linear templates.
https://www.lead-dbs.org/about-the-mni-spaces/
from IPython.display import Image
Image(filename = "./assets/atlases.png", width=1000, height=800)
# import warnings
# warnings.filterwarnings('ignore')
# import nilearn
# from nilearn import plotting
# from nilearn import image
# from nilearn.datasets import load_mni152_template
# import nibabel as nib
# %matplotlib inline
You can think of the image affine as a combination of a series of transformations to go from voxel coordinates to mm coordinates in terms of the magnet isocenter. Here is the EPI affine broken down into a series of transformations, with the results shown on the localizer image:
Image(filename = "./assets/affine_1.png", width=200, height=100)
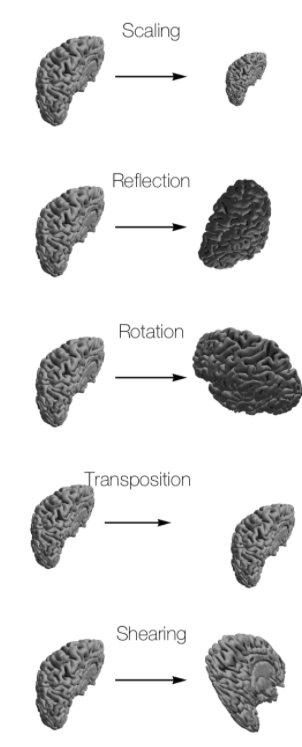
Affine transformations and rigid transformations#
Why affine qform and sform tranformations are important? sform allows full 12 parameter affine transfrom to be encoded, however qform 9 parameter is limited to encoding translations, rotations (via a quaternion representation) and isotropic zooms.
https://www.lead-dbs.org/about-the-mni-spaces/
# !wget --progress=bar:force:noscroll -P /tmp http://www.bic.mni.mcgill.ca/~vfonov/icbm/2009/mni_icbm152_nlin_sym_09a_nifti.zip && \
# mkdir -p /workspace/data/mni_icbm152_nlin_sym_09a && \
# unzip -o -d /workspace/data/tmp/mni_icbm152_nlin_sym_09a_nifti.zip && \
# rm -r /tmp/mni_icbm152_nlin_sym_09a_nifti.zip
# nii_file = nib.load('/workspace/data/raw/100206/unprocessed/3T/T1w_MPR1/100206_3T_T1w_MPR1.nii.gz')
# # Load ing MNI template
# mni_space_template = nib.load('/workspace/data/mni_icbm152_nlin_sym_09a/mni_icbm152_t1_tal_nlin_sym_09a.nii') # Full template
# nilearn.plotting.plot_anat(nii_file,
# title='Nifti T1 Native Space', annotate=True)
# nilearn.plotting.plot_anat(mni_space_template,
# title='Nifti MNI152 Space')
# diff = nilearn.plotting.plot_anat(nii_file,
# title='Space displacement')
# diff.add_contours(mni_space_template, threshold=70)
Let’s check how using qform we can translate images.
First: Check images orientation between MNI template and out T1 image.
print(“””Orintation comparison before transform:
T1 native image orientation:\n {0}
MNI Space template brain:\n {1} “””.format(nii_file.affine, mni_space_template.affine))
Second: Check shapes of image
# print("""Shape comparison before transform:
# - T1 native image affine:\n {0}
# - MNI Space template brain:\n {1}
# """.format(nii_file.shape, mni_space_template.shape))
Third: use from_matvec method, to create 4x4 matrix from 3x3 vector to add translation of translate to dot with qform header
# from nibabel.affines import from_matvec, to_matvec, apply_affine
# import numpy as np
# import matplotlib.pyplot as plt
# # reflect = np.array([[-1, 0, 0],
# # [0, 1, 0],
# # [0, 0, 1]])
# reflect = np.array([[1, 0, 0],
# [0, 1, 0],
# [0, 0, 1]])
# translate_affine = from_matvec(reflect, [50, 0, 0]) # shift 50 x direction
# translate_affine
# transformed = np.dot(nii_file.get_qform(), translate_affine)
# #nii_file.affine = transformed
# print(transformed)
# print(nii_file.get_qform())
Finally, create new nifti with same array if intensities but with new qform header after translation with NIFTI1Image with new
# header_info = nii_file.header
# vox_data = np.array(nii_file.get_fdata())
# transformed_nii = nib.Nifti1Image(vox_data, transformed)
# tranfsorm_image = nilearn.plotting.plot_anat(transformed_nii,
# title='Space displacement', annotate=True)
# tranfsorm_image.add_overlay(nii_file, alpha=0.5)
# transform_image = nilearn.plotting.plot_anat(transformed_nii,
# title='Space displacement', annotate=True)
# transform_image.add_overlay(nii_file,threshold=0.8e3, colorbar=True)
Try: How about rotation?
# cos_gamma = np.cos(0.3)
# sin_gamma = np.sin(0.3)
# rotation_affine = np.array([[1, 0, 0, 0],
# [0, cos_gamma, -sin_gamma, 0],
# [0, sin_gamma, cos_gamma, 0],
# [0, 0, 0, 1]])
# rotation_affine
# after_rot = image.resample_img(nii_file,
# target_affine=rotation_affine.dot(nii_file.affine),
# target_shape=nii_file.shape,
# interpolation='continuous')
# chimera = nib.Nifti1Image(after_rot.get_fdata(), nii_file.affine) # get_data - error
# tranfsorm_image = nilearn.plotting.plot_anat(nii_file,
# title='Rotation', annotate=True)
# tranfsorm_image.add_overlay(chimera, alpha=0.5)
# after_rot.shape == nii_file.shape
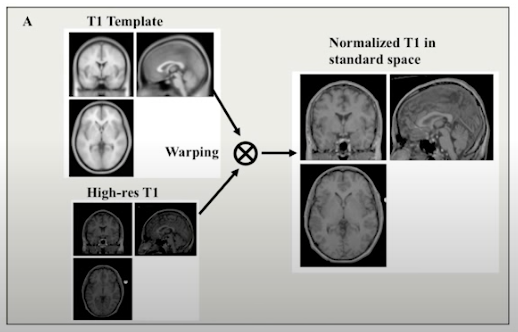
Spatial Normalization Methods#
All brains are different. The brain size of two subject can differ in size by up to 30%.
There may also be substantial variation in the shapes of the brain.
Normalization allows one to stretch, squeeze and warp each brain so that it is the same as some standard brain.
Pros/cons:
results can be generalized to larger population;
results can be compared across studies;
results can be averaged across subjects;
potential errors (always make visual control of results);
reduces spatial resolution
Image(filename = "./assets/normalization.png", width=400, height=100)
ANTs Registration#
Volume-based registration method, often used for corregistration between series. \
https://nipype.readthedocs.io/en/1.1.7/interfaces/generated/interfaces.ants/registration.html
ANTsX/ANTs
# from nipype.interfaces.ants import RegistrationSynQuick
# reg = RegistrationSynQuick()
# reg.inputs.fixed_image = '/workspace/data/mni_icbm152_nlin_sym_09a/mni_icbm152_t1_tal_nlin_sym_09a.nii'
# reg.inputs.moving_image = '/workspace/data/raw/100206/unprocessed/3T/T1w_MPR1/100206_3T_T1w_MPR1.nii.gz'
# reg.inputs.output_prefix = 'subject_name'
# print(reg.cmdline)
# reg.run()
ANTs initialize + ANTs transform#
Next methods of ants compute optimal transformation and produces ./mat transformation matrix, then calling registation() to apply it.
ANTs initialize affine between two spaces and outputs transformation matrix ANTs Transform takes affine matrix and output target space image
# from nipype.interfaces.ants import AffineInitializer
# init = AffineInitializer()
# init.inputs.fixed_image = '/workspace/data/mni_icbm152_nlin_sym_09a/mni_icbm152_t1_tal_nlin_sym_09a.nii'
# init.inputs.moving_image = '/workspace/data/raw/100206/unprocessed/3T/T1w_MPR1/100206_3T_T1w_MPR1.nii.gz'
# init.inputs.out_file = '/workspace/data/transfm.mat'
# print(init.cmdline)
# init.run()
Apply a transform list to map an image from one domain to another.
# from nipype.interfaces.ants import ApplyTransforms
# at = ApplyTransforms()
# at.inputs.input_image = '/workspace/data/raw/100206/unprocessed/3T/T1w_MPR1/100206_3T_T1w_MPR1.nii.gz'
# at.inputs.reference_image = '/workspace/data/mni_icbm152_nlin_sym_09a/mni_icbm152_t1_tal_nlin_sym_09a.nii'
# at.inputs.transforms = '/workspace/data/transfm.mat'
# at.inputs.output_image = './sub-100206_MNI159_space.nii.gz'
# at.inputs.interpolation = 'Linear'
# print(at.cmdline)
# at.run()
Compare shape and orientation done by two methods
# nii_file_mni152_antsreg = nib.load('./sub-100206_MNI159_space.nii.gz')
# nii_file_mni152_antstransform = nib.load('./subject_nameWarped.nii.gz')
# print("""Orientation comparison after transform:
# - T1 native after transform:\n {0}
# - MNI Space template:\n {1}
# - T1 native after transform with affine: \n {2}
# """.format(nii_file_mni152_antsreg.affine, mni_space_template.affine, nii_file_mni152_antstransform.affine))
# print("""Shape comparison after transform:
# - T1 native after transform:\n {0}
# - MNI Space template brain:\n {1}
# - T1 native after transform with antstransform: \n {2}
# """.format(nii_file_mni152_antsreg.shape, mni_space_template.shape, nii_file_mni152_antstransform.shape))
T1w and template alignment
# antstransform_result = nilearn.plotting.plot_anat(nii_file_mni152_antsreg)
# antstransform_result.add_contours(mni_space_template, threshold=70, title='Nifti MNI152 Space')
# import nilearn
# from nilearn import plotting
# nilearn.plotting.view_img(nii_file, bg_img=mni_space_template, threshold='auto')
Tips:
Run a bias correction before antsRegistration (i.e. N4). It helps getting better registration. \
Remove the skull before antsRegistration. If you have two brain-only images, you can be sure that surrounding tissues (i.e. the skull) will not take a toll on the registration accuracy. If you are using these skull-stripped versions, you can avoid using the mask, because you want the registration to use the “edge” features. If you use a mask, anything out of the mask will not be considered, the algorithm will try to match what’s inside the brain, but not the edge of the brain itself (see Nick’s explanation here). \
Never register a lesioned brain with a healthy brain without a proper mask. The algorithm will just pull the remaining parts of the lesioned brain to fill “the gap”. Despite initial statements that you can register lesioned brains without the need to mask out the lesion, there is evidence showing that results without lesion masking are sub-optimal. If you really don’t have the lesion mask, even a coarse and imprecise drawing of lesions helps (see Andersen 2010). \
Don’t forget to read the parts of the manual (ANTsX/ANTs) related to registration.
File formats convertation - volume-to-volume#
You can convert file from preprocessing freesurfer to mni and t1 spaces, using antsTransform with fresurfer native command: mri_convert
# !mri_convert --in_type mgz --out_type nii --out_orientation RAS \
# /workspace/data/freesurfer_preproc/100206/mri/T1.mgz \
# /workspace/T1_fs_preprocessed.nii
# freesurfer_t1 = nib.load('./T1_fs_preprocessed.nii')
# freesurfer_t1.shape
# freesurfer_t1.affine
# freesurfer_plot = nilearn.plotting.plot_anat(freesurfer_t1)
# freesurfer_plot.add_contours(mni_space_template)
# import os
# Implement ANTs registration to allign mni and freesurfer template
# PUT YOUR CODE HERE
# reg = RegistrationSynQuick()
# reg.inputs.moving_image = os.path.abspath('./T1_fs_preprocessed.nii')
# reg.inputs.fixed_image = os.path.abspath('...')
# ...
# reg = RegistrationSynQuick()
# reg.inputs.moving_image = os.path.abspath('./T1_fs_preprocessed.nii')
# reg.inputs.fixed_image = os.path.abspath('/workspace/data/mni_icbm152_nlin_sym_09a/mni_icbm152_t1_tal_nlin_sym_09a.nii')
# reg.inputs.output_prefix = 'fs'
# print(reg.cmdline)
# reg.run()
# nilearn.plotting.plot_img('fsInverseWarped.nii.gz', bg_img='fsWarped.nii.gz', threshold='auto', alpha=0.8)
